Single-cell RNA-seq Analysis of Zebrafish Heart
Cite: If you use this website, it would be very grateful if you could please cite: J Koth, X Wang and AC Killen et al.
Code: Scripts used to analysis this dataset is available at https://github.com/SharonWang/Zebrafish_runx1_analysis
Main content of the website:
UMAP: UMAP visualisation of the data
Violinplots: Violin plots of the selected gene
GeneAnno: Differential expression results and pathway analysis of gene lists
Study Design:
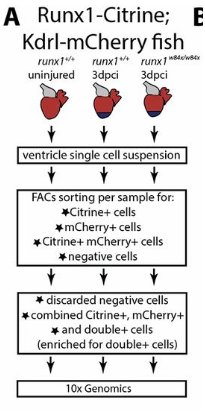
Samples:
Main visualisation:
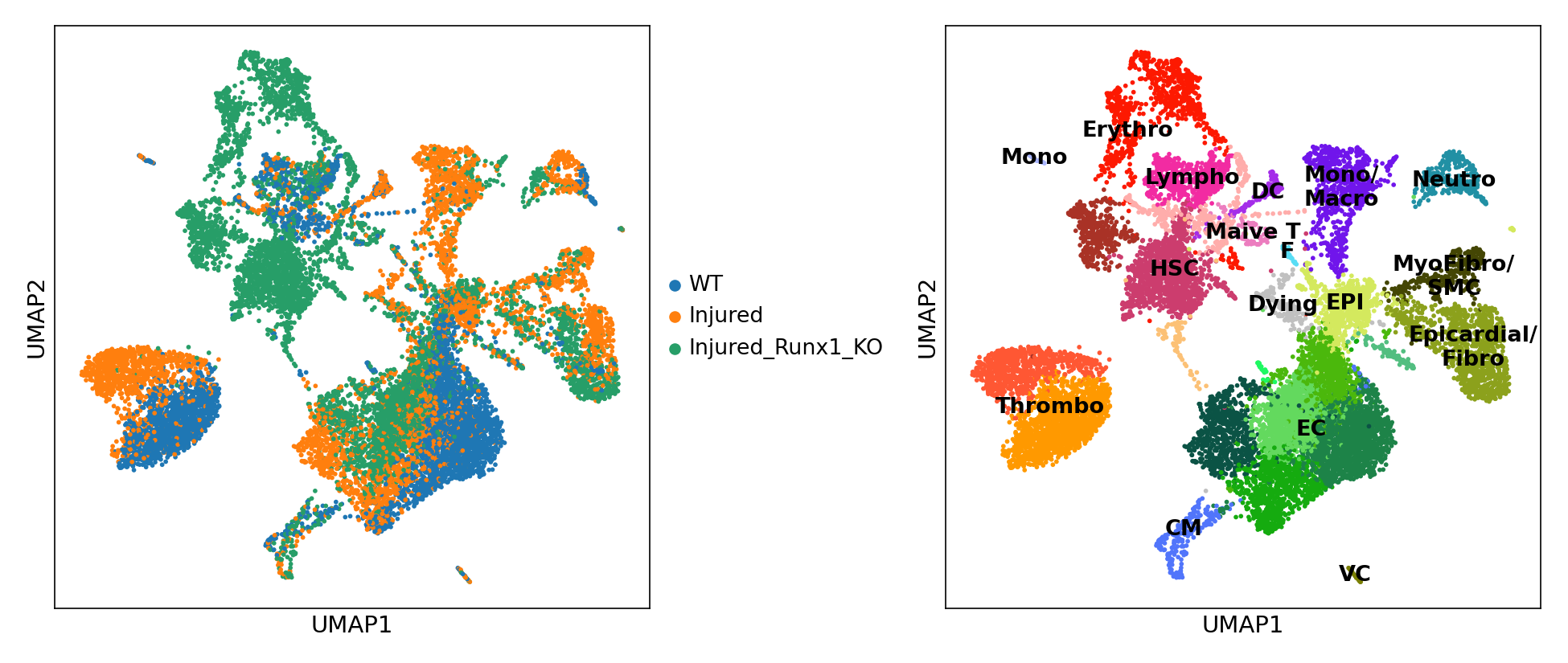
EC: Endothelial/Endocardial cells;
EPI: Epicardial cells;
CM: Cardiomyocytes;
VC: Valve cells;
MyoFibro: Myofibroblasts;
Fibro: Fibroblasts;
Neutro: Neutrophils;
Mono: Monocytes;
Macro: Macrophages;
DC: Dendritic cells;
Naive T: Naive T cells;
HSC: Haematopoietic stem cells;
Lympho: Lymphocytes;
Erythro: Erythrocytes;
Thrombo: Thrombocytes;
Violinplot
Please click on a gene in the differential expression result table to plot the gene expression on UMAP